Last week I released an update to the BC Leps Curated List which updates the BC list (now at 2855) and adds New Additions and Taxon Changes sections. This has been a bit of a labour of love these last few months, with the help and advice from a couple of members of the BC Leps team who’ve worked on publishing and tracking the checklist for many years.
The idea to this tool was to automate the generation of a checklist of lepidoptera for British Columbia. The checklist would be a “live” list that reflects our current knowledge of the species in BC, rather than requiring people to rely on data that was published every few years, thus out of date. But broader than that, I wanted to create a script that can be used for any taxon and any place. In the era of iNat and citizen science projects, the wealth of data flooding in is staggering: keeping your head above water parsing the data for legitimate additions to your list is tough!
New Additions
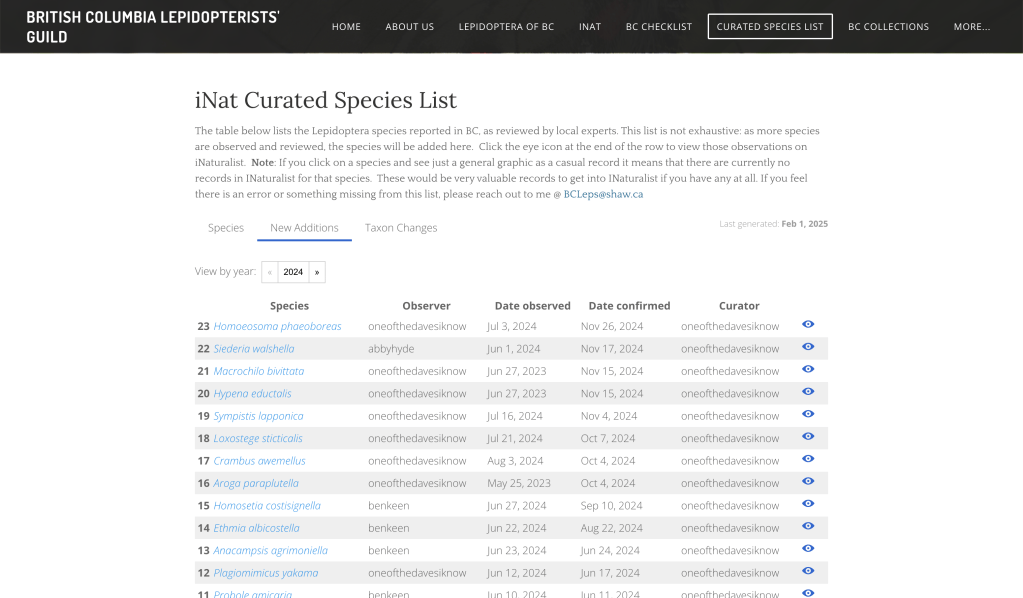
Prior to this update, it was virtually impossible to identify exactly which were the new species added to our checklist over time. Such useful information felt conspicuously absent.
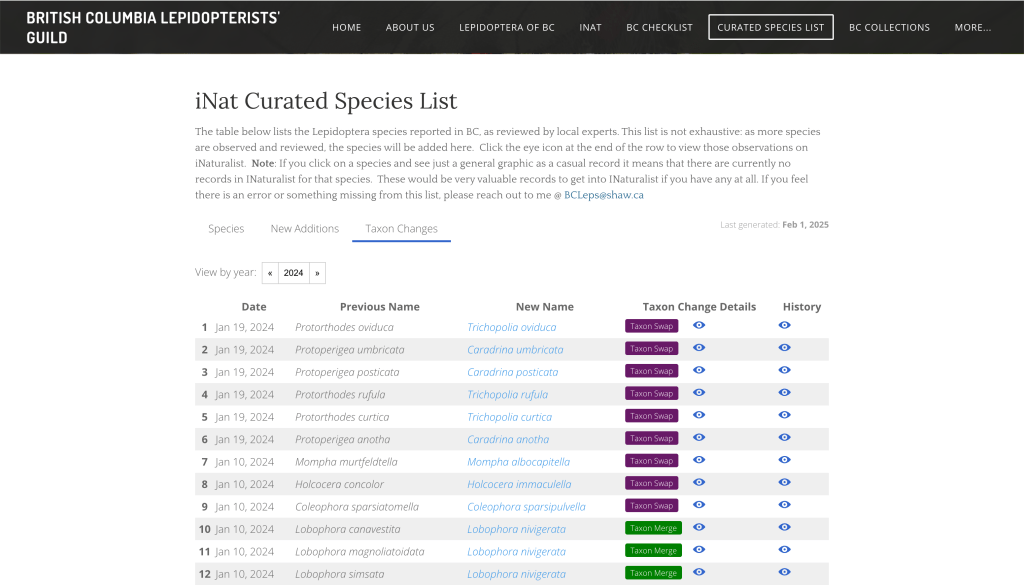
Taxon Changes
While working on the New Additions, I noticed that the data I was parsing also included taxon change data. Well, well, well! This was another area I that’d be useful to expose. I always seem to be a step behind with taxonomy shifts, so showing a summary of the changes with links back to more information on iNat would be a nice addition.
So… is the project finally done?
Nuh-uh! There’s still a big question mark hanging over our usage of iNat to store the baseline data. Early on, we fed into iNat “casual” observations of certain species known to be in the province but don’t yet have iNat observations in BC. Frankly I was under the impression that this was just fine: I’ve put in the occasional “casual” observation without any associated photo because I knew I saw that species, and that’s considered well within acceptable use for iNat. But the concern has been raised that perhaps it’s not valid in our case, since it’s not an individual making those observations, but on behalf of a group. That’s a legit concern, so we may well need to work around it.
The benefit to housing the data in iNat is that we benefit from the larger iNat community’s maintenance of taxon changes. INat Curator Bob logs a taxon split for species X here; iNat Curator Jim logs a different taxon merge for another taxon over there. By housing the baseline data in iNat, we automatically inherit those changes; we simply re-run our script to regenerate our checklist. That’s it! All automated. If we have to delete the baseline observation data, we’d have to find a way to ensure our checklist’s taxonomy was always up to date. Hmph!
I have a technical solution in mind, which I believe is the only way forward: we move the baseline data to our own code, then every time we ran the script it would identify which taxons are no longer current, then require a person to review the taxon changes and manually update our baseline data. Ideal? No – but I think it’s perfectly feasible and realistic. There aren’t that many taxon shifts per year, so a chore task we have to perform every couple of months doesn’t seem too bad.
Still, more internal discussions to be had first. But ultimately I’m anxious to speak to the iNat team about our options. This is a really neat tool and I’d love to see it adopted beyond our single use case. And if we can make the tool as simple as possible to use, it’ll lower the barrier to entry for everyone.